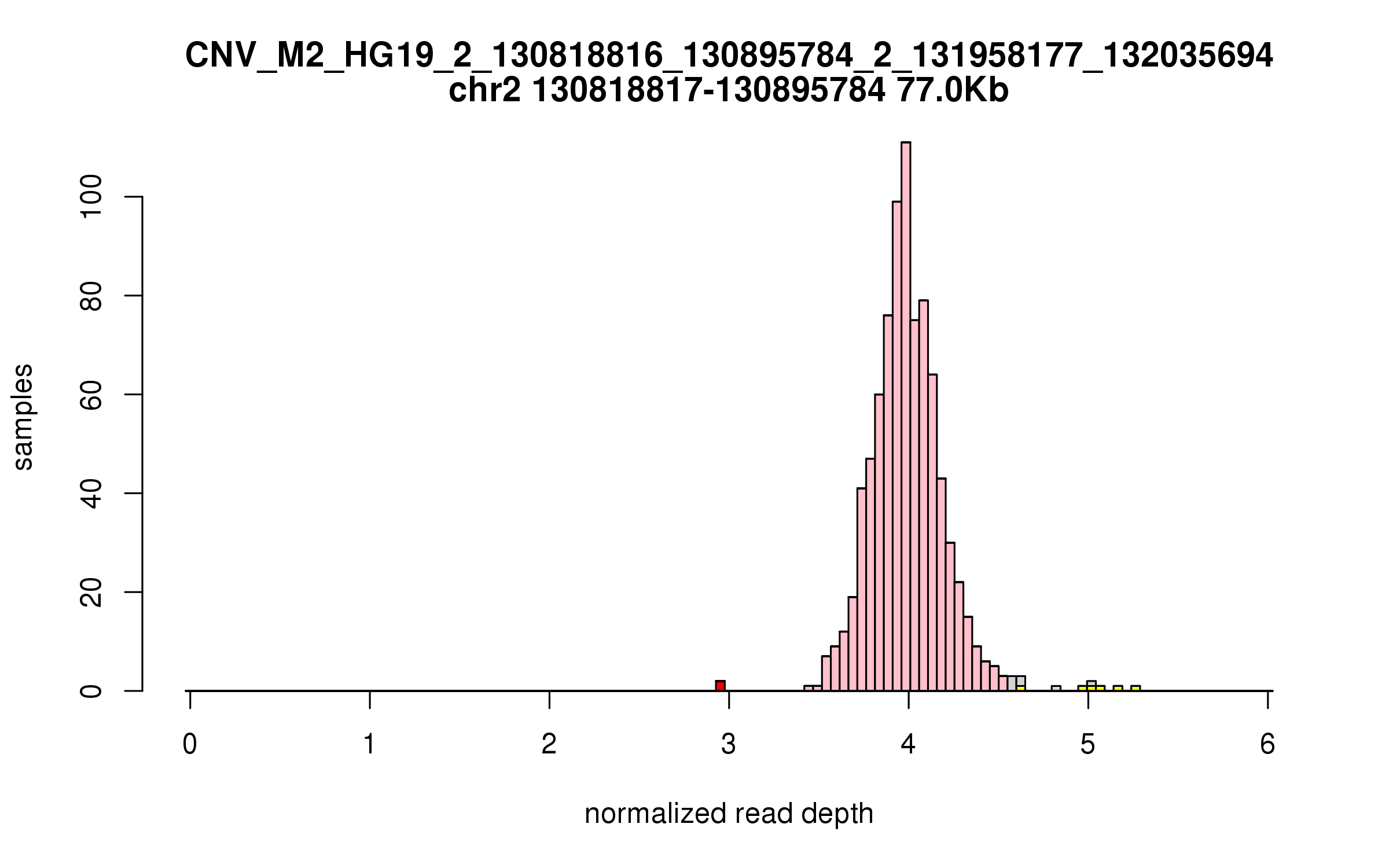
Variant CNV_M2_HG19_2_130818816_130895784_2_131958177_132035694
Location: chr2:130818816-130895784 (76Kb), chr2:131958177-132035694 (77Kb), distance: 1.1Mb
Site classification: Multi-allelic
Variant DCN frequency: 0.009
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.009
Genes Overlapped
| PLEKHB2 | INTRON | NA |
| POTEE | GENE | POTE ankyrin domain family member E |
| POTEF | GENE | POTE ankyrin domain family member F |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 0 0 45 0
CEU 0 0 0 0 66 0
CHB 0 0 0 0 77 1
CHS 0 0 0 0 89 2
CLM 0 0 0 1 48 0
FIN 0 0 0 0 62 0
GBR 0 0 0 1 53 0
IBS 0 0 0 0 6 0
JPT 0 0 0 0 68 1
LWK 0 0 0 0 67 2
MXL 0 0 0 0 54 0
PUR 0 0 0 0 50 2
TSI 0 0 0 0 83 0
YRI 0 0 0 0 71 0
AFR 0 0 0 0 138 2
AFR+ 0 0 0 0 183 2
AMR 0 0 0 1 152 2
ASN 0 0 0 0 234 4
EUR 0 0 0 1 270 0
ALL 0 0 0 2 839 8
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 0 0 45 0
CEU 0 0 0 0 66 0
CHB 0 0 0 0 77 1
CHS 0 0 0 0 89 0
CLM 0 0 0 1 48 0
FIN 0 0 0 0 62 0
GBR 0 0 0 1 53 0
IBS 0 0 0 0 6 0
JPT 0 0 0 0 68 1
LWK 0 0 0 0 67 2
MXL 0 0 0 0 54 0
PUR 0 0 0 0 50 2
TSI 0 0 0 0 78 0
YRI 0 0 0 0 71 0
AFR 0 0 0 0 138 2
AFR+ 0 0 0 0 183 2
AMR 0 0 0 1 152 2
ASN 0 0 0 0 234 2
EUR 0 0 0 1 265 0
ALL 0 0 0 2 834 6
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI