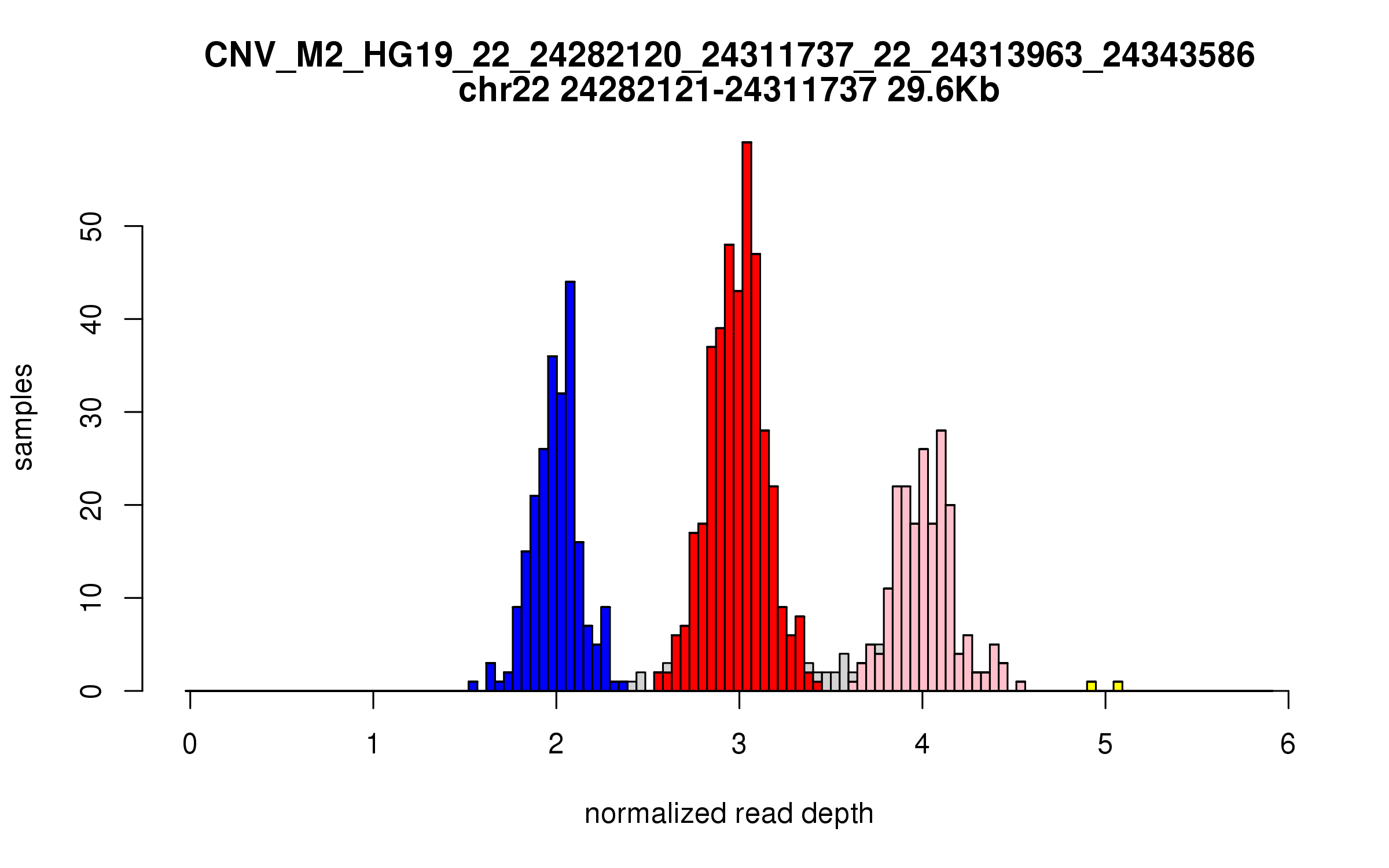
Variant CNV_M2_HG19_22_24282120_24311737_22_24313963_24343586
Location: chr22:24282120-24311737 (29Kb), chr22:24313963-24343586 (29Kb), distance: 2.2Kb
Site classification: Multi-allelic
Variant DCN frequency: 0.519
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.519
Genes Overlapped
| DDT | CDS | D-dopachrome decarboxylase |
| DDTL | CDS | D-dopachrome decarboxylase-like protein |
| DDTL | EXON | D-dopachrome decarboxylase-like protein |
| GSTT2 | GENE | glutathione S-transferase theta 2 |
| GSTT2B | GENE | glutathione S-transferase theta-2B |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 6 24 15 0
CEU 0 0 32 27 7 0
CHB 0 0 16 43 19 0
CHS 0 0 22 40 29 0
CLM 0 0 12 23 13 1
FIN 0 0 25 31 6 0
GBR 0 0 22 20 12 0
IBS 0 0 1 4 1 0
JPT 0 0 16 36 17 0
LWK 0 0 10 35 23 1
MXL 0 0 17 22 15 0
PUR 0 0 9 29 14 0
TSI 0 0 33 35 15 0
YRI 0 0 9 39 23 0
AFR 0 0 19 74 46 1
AFR+ 0 0 25 98 61 1
AMR 0 0 38 74 42 1
ASN 0 0 54 119 65 0
EUR 0 0 113 117 41 0
ALL 0 0 230 408 209 2
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 6 24 15 0
CEU 0 0 32 27 7 0
CHB 0 0 16 43 19 0
CHS 0 0 22 38 27 0
CLM 0 0 12 21 11 1
FIN 0 0 25 29 6 0
GBR 0 0 22 20 12 0
IBS 0 0 1 3 1 0
JPT 0 0 16 36 17 0
LWK 0 0 10 35 23 1
MXL 0 0 16 22 14 0
PUR 0 0 9 29 11 0
TSI 0 0 33 35 15 0
YRI 0 0 9 39 23 0
AFR 0 0 19 74 46 1
AFR+ 0 0 25 98 61 1
AMR 0 0 37 72 36 1
ASN 0 0 54 117 63 0
EUR 0 0 113 114 41 0
ALL 0 0 229 401 201 2
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI