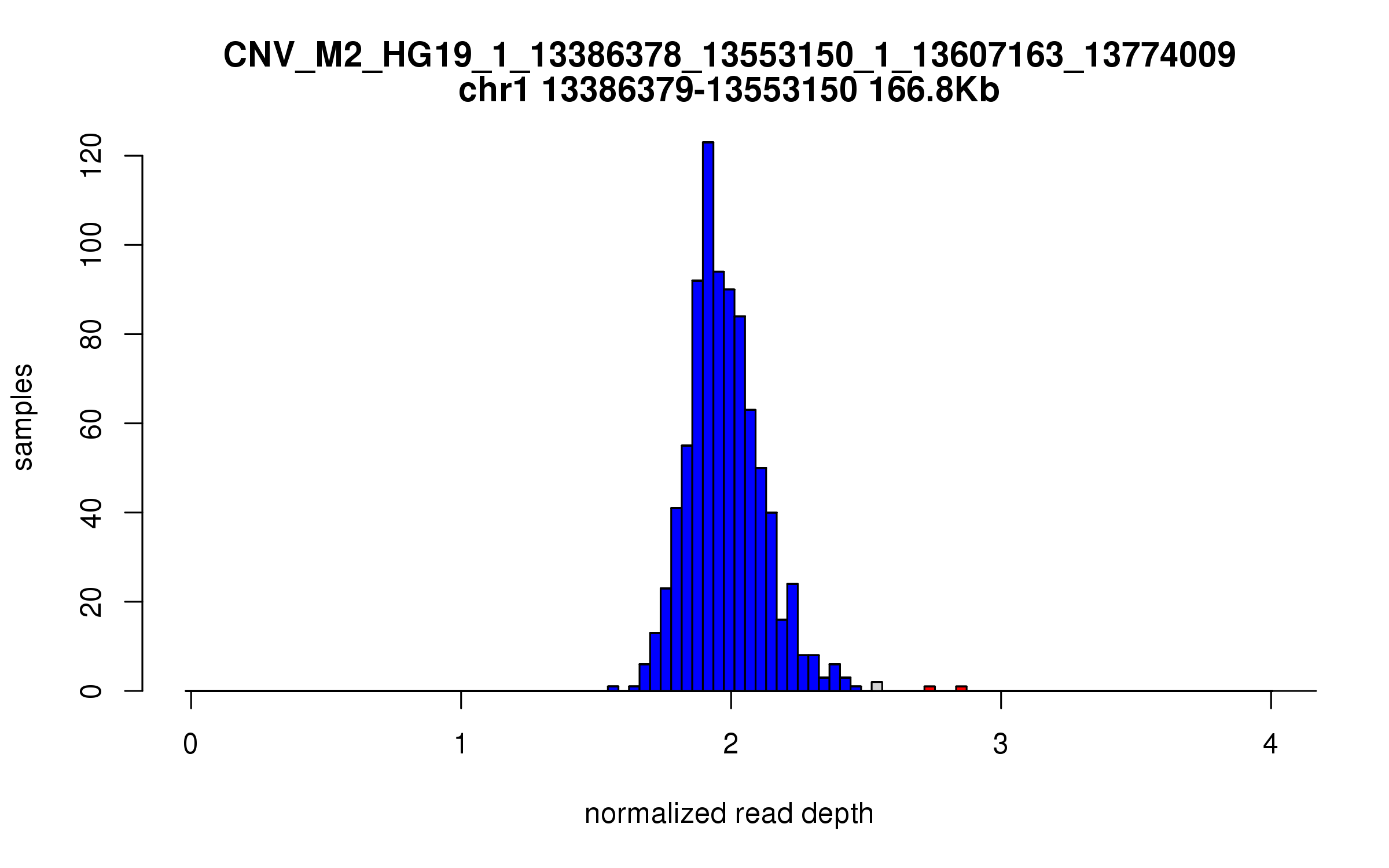
Variant CNV_M2_HG19_1_13386378_13553150_1_13607163_13774009
Location: chr1:13386378-13553150 (166Kb), chr1:13607163-13774009 (166Kb), distance: 54Kb
Site classification: Duplication
Variant DCN frequency: 0.002
Genes Overlapped
Site classification: Duplication
Variant DCN frequency: 0.002
Genes Overlapped
| PRAMEF13 | GENE | PRAME family member 13 |
| PRAMEF14 | GENE | NA |
| PRAMEF15 | GENE | PRAME family member 9/15 |
| PRAMEF16 | GENE | PRAME family member 16 |
| PRAMEF17 | GENE | PRAME family member 17 |
| PRAMEF18 | GENE | PRAME family member 18 |
| PRAMEF19 | GENE | PRAME family member 19 |
| PRAMEF20 | GENE | PRAME family member 20/21 |
| PRAMEF21 | GENE | PRAME family member 20/21 |
| PRAMEF8 | GENE | PRAME family member 8 |
| PRAMEF9 | GENE | PRAME family member 9/15 |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3
ASW 0 0 45 0
CEU 0 0 65 1
CHB 0 0 78 0
CHS 0 0 91 0
CLM 0 0 49 0
FIN 0 0 62 0
GBR 0 0 54 0
IBS 0 0 6 0
JPT 0 0 69 0
LWK 0 0 69 0
MXL 0 0 54 0
PUR 0 0 52 0
TSI 0 0 82 1
YRI 0 0 70 1
AFR 0 0 139 1
AFR+ 0 0 184 1
AMR 0 0 155 0
ASN 0 0 238 0
EUR 0 0 269 2
ALL 0 0 846 3
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3
ASW 0 0 45 0
CEU 0 0 65 1
CHB 0 0 78 0
CHS 0 0 91 0
CLM 0 0 49 0
FIN 0 0 62 0
GBR 0 0 54 0
IBS 0 0 6 0
JPT 0 0 69 0
LWK 0 0 69 0
MXL 0 0 54 0
PUR 0 0 52 0
TSI 0 0 82 1
YRI 0 0 69 0
AFR 0 0 138 0
AFR+ 0 0 183 0
AMR 0 0 155 0
ASN 0 0 238 0
EUR 0 0 269 2
ALL 0 0 845 2
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI