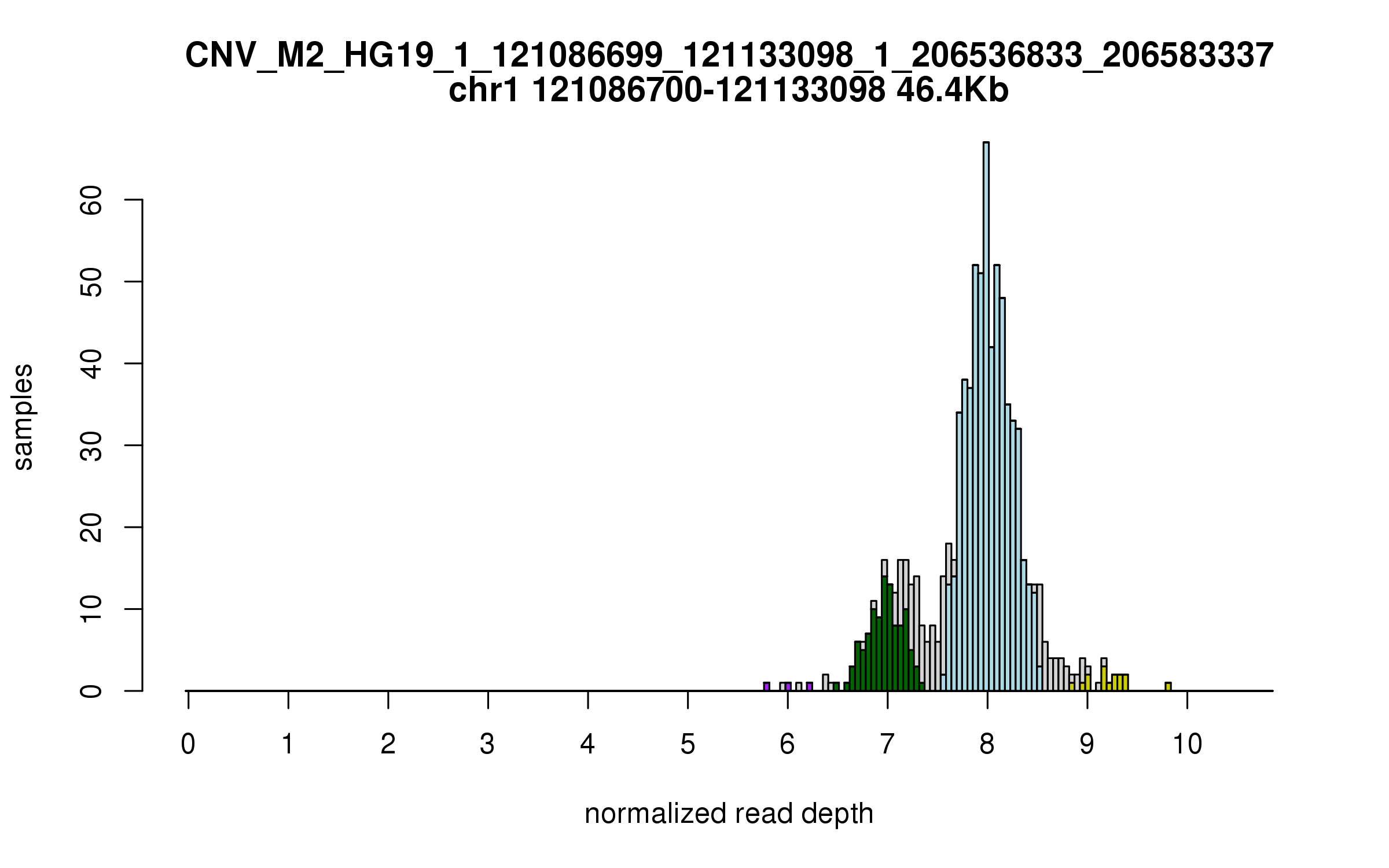
Variant CNV_M2_HG19_1_121086699_121133098_1_206536833_206583337
Location: chr1:121086699-121133098 (46Kb), chr1:206536833-206583337 (46Kb), distance: 85Mb
Site classification: Multi-allelic
Variant DCN frequency: 0.169
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.169
Genes Overlapped
| SRGAP2 | CDS | SLIT-ROBO Rho GTPase-activating protein 2 |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5 CN6 CN7 CN8 CN9
ASW 0 0 0 0 0 0 0 2 40 3
CEU 0 0 0 0 0 0 1 16 49 0
CHB 0 0 0 0 0 0 0 19 57 2
CHS 0 0 0 0 0 0 0 16 70 5
CLM 0 0 0 0 0 0 0 9 38 2
FIN 0 0 0 0 0 0 0 13 46 3
GBR 0 0 0 0 0 0 0 6 45 3
IBS 0 0 0 0 0 0 0 0 6 0
JPT 0 0 0 0 0 0 0 16 51 2
LWK 0 0 0 0 0 0 0 7 62 0
MXL 0 0 0 0 0 0 1 7 43 3
PUR 0 0 0 0 0 0 0 15 37 0
TSI 0 0 0 0 0 0 1 18 63 1
YRI 0 0 0 0 0 0 1 6 63 1
AFR 0 0 0 0 0 0 1 13 125 1
AFR+ 0 0 0 0 0 0 1 15 165 4
AMR 0 0 0 0 0 0 1 31 118 5
ASN 0 0 0 0 0 0 0 51 178 9
EUR 0 0 0 0 0 0 2 53 209 7
ALL 0 0 0 0 0 0 4 150 670 25
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5 CN6 CN7 CN8 CN9
ASW 0 0 0 0 0 0 0 1 37 3
CEU 0 0 0 0 0 0 1 14 40 0
CHB 0 0 0 0 0 0 0 18 55 1
CHS 0 0 0 0 0 0 0 4 60 2
CLM 0 0 0 0 0 0 0 5 35 0
FIN 0 0 0 0 0 0 0 6 36 3
GBR 0 0 0 0 0 0 0 2 32 1
IBS 0 0 0 0 0 0 0 0 6 0
JPT 0 0 0 0 0 0 0 12 51 1
LWK 0 0 0 0 0 0 0 3 58 0
MXL 0 0 0 0 0 0 0 6 39 2
PUR 0 0 0 0 0 0 0 10 33 0
TSI 0 0 0 0 0 0 1 17 52 1
YRI 0 0 0 0 0 0 1 5 60 1
AFR 0 0 0 0 0 0 1 8 118 1
AFR+ 0 0 0 0 0 0 1 9 155 4
AMR 0 0 0 0 0 0 0 21 107 2
ASN 0 0 0 0 0 0 0 34 166 4
EUR 0 0 0 0 0 0 2 39 166 5
ALL 0 0 0 0 0 0 3 103 594 15
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI