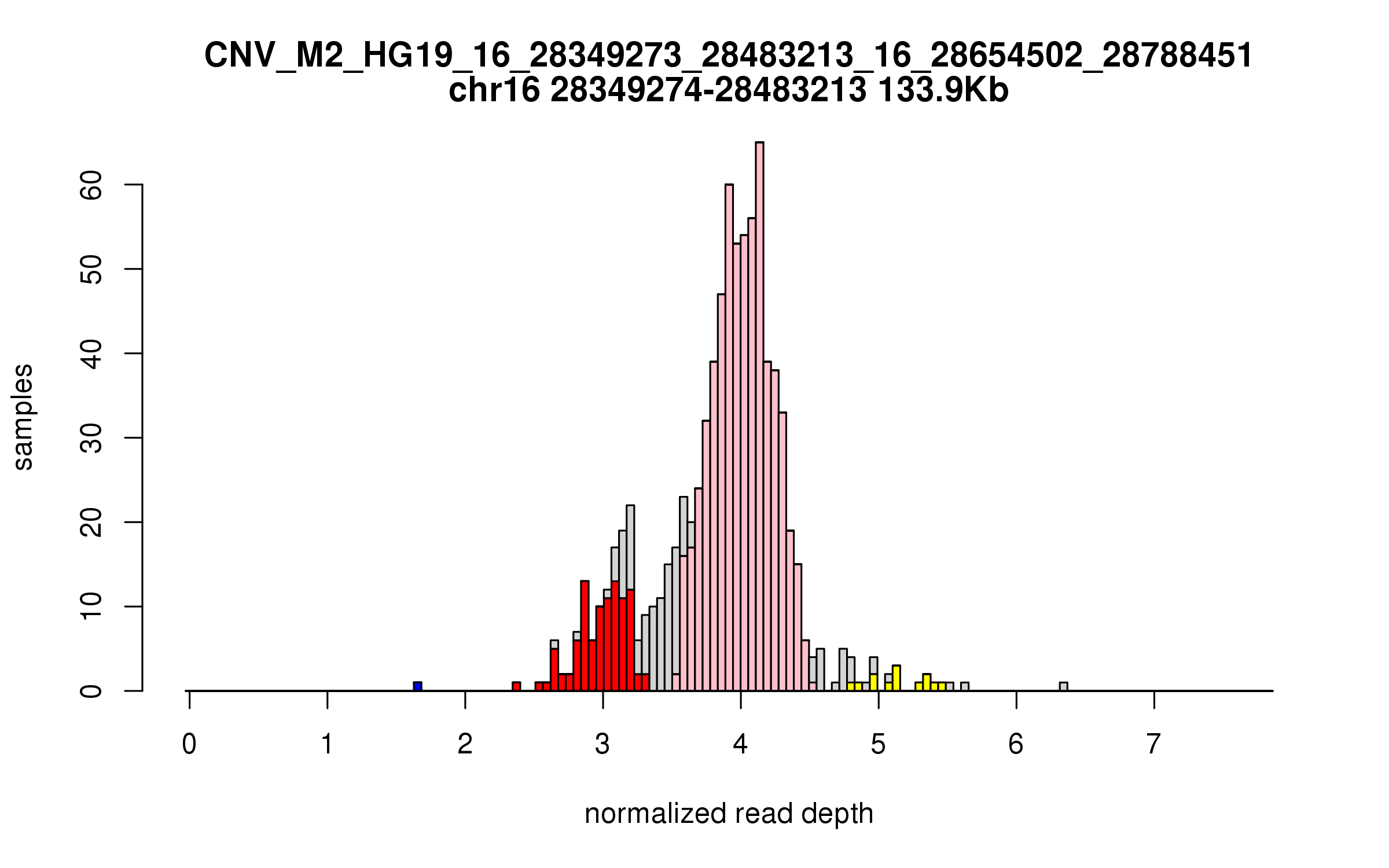
Variant CNV_M2_HG19_16_28349273_28483213_16_28654502_28788451
Location: chr16:28349273-28483213 (133Kb), chr16:28654502-28788451 (133Kb), distance: 171Kb
Site classification: Multi-allelic
Variant DCN frequency: 0.153
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.153
Genes Overlapped
| A-575C2.4 | CDS | NA |
| A-575C2.4 | GENE | NA |
| CLN3 | EXON | Cln3p |
| EIF3C | GENE | eukaryotic translation initiation factor 3 subunit C |
| EIF3CL | GENE | eukaryotic translation initiation factor 3 subunit C |
| NPIPL1 | GENE | NA |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5 CN6
ASW 0 0 0 6 37 1 1
CEU 0 0 0 8 54 4 0
CHB 0 0 0 15 59 4 0
CHS 0 0 1 24 66 0 0
CLM 0 0 0 9 40 0 0
FIN 0 0 0 14 47 1 0
GBR 0 0 0 6 47 1 0
IBS 0 0 0 0 6 0 0
JPT 0 0 0 11 55 3 0
LWK 0 0 0 1 65 3 0
MXL 0 0 0 9 43 2 0
PUR 0 0 0 7 43 2 0
TSI 0 0 0 19 63 1 0
YRI 0 0 0 5 63 3 0
AFR 0 0 0 6 128 6 0
AFR+ 0 0 0 12 165 7 1
AMR 0 0 0 25 126 4 0
ASN 0 0 1 50 180 7 0
EUR 0 0 0 47 217 7 0
ALL 0 0 1 134 688 25 1
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 0 5 36 1
CEU 0 0 0 5 51 3
CHB 0 0 0 12 56 1
CHS 0 0 1 19 49 0
CLM 0 0 0 6 32 0
FIN 0 0 0 7 41 1
GBR 0 0 0 1 44 0
IBS 0 0 0 0 5 0
JPT 0 0 0 11 53 2
LWK 0 0 0 1 55 1
MXL 0 0 0 5 41 2
PUR 0 0 0 5 30 0
TSI 0 0 0 16 59 1
YRI 0 0 0 4 63 1
AFR 0 0 0 5 118 2
AFR+ 0 0 0 10 154 3
AMR 0 0 0 16 103 2
ASN 0 0 1 42 158 3
EUR 0 0 0 29 200 5
ALL 0 0 1 97 615 13
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI