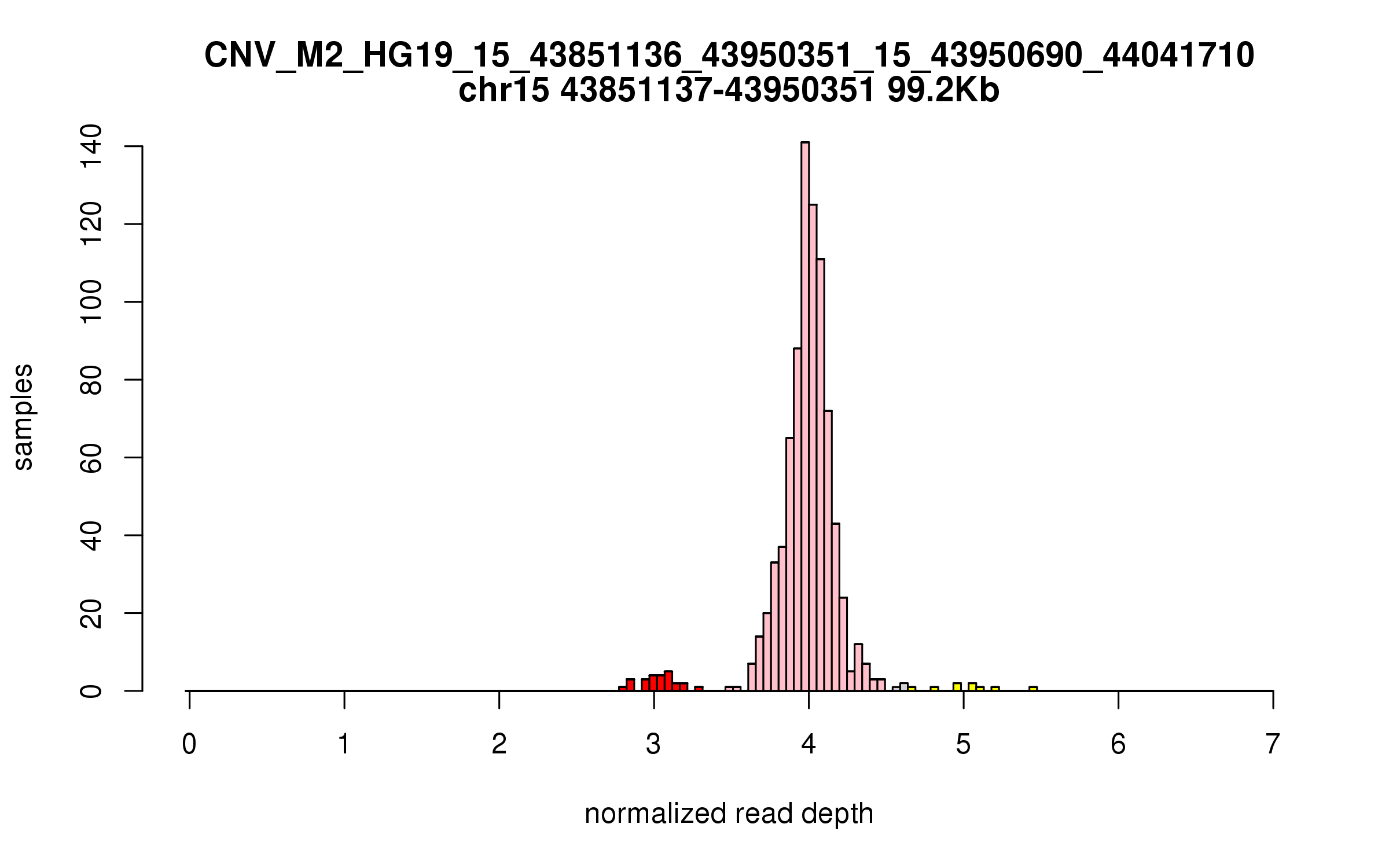
Variant CNV_M2_HG19_15_43851136_43950351_15_43950690_44041710
Location: chr15:43851136-43950351 (99Kb), chr15:43950690-44041710 (91Kb), distance: 338b
Site classification: Multi-allelic
Variant DCN frequency: 0.040
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.040
Genes Overlapped
| CATSPER2 | CDS | cation channel sperm-associated protein 2 |
| CATSPER2 | EXON | cation channel sperm-associated protein 2 |
| CKMT1A | GENE | creatine kinase U-type, mitochondrial |
| CKMT1B | GENE | creatine kinase U-type, mitochondrial precursor |
| PDIA3 | CDS | protein disulfide-isomerase A3 precursor |
| PPIP5K1 | CDS | inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 1 |
| STRC | CDS | stereocilin precursor |
| STRC | EXON | stereocilin precursor |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 0 1 44 0
CEU 0 0 0 2 64 0
CHB 0 0 0 2 76 0
CHS 0 0 0 3 88 0
CLM 0 0 0 2 43 4
FIN 0 0 0 3 59 0
GBR 0 0 0 4 49 1
IBS 0 0 0 0 6 0
JPT 0 0 0 1 66 2
LWK 0 0 0 2 66 1
MXL 0 0 0 0 54 0
PUR 0 0 0 1 50 1
TSI 0 0 0 1 81 1
YRI 0 0 0 3 67 1
AFR 0 0 0 5 133 2
AFR+ 0 0 0 6 177 2
AMR 0 0 0 3 147 5
ASN 0 0 0 6 230 2
EUR 0 0 0 10 259 2
ALL 0 0 0 25 813 11
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 0 1 44 0
CEU 0 0 0 2 64 0
CHB 0 0 0 2 76 0
CHS 0 0 0 3 88 0
CLM 0 0 0 2 43 3
FIN 0 0 0 3 59 0
GBR 0 0 0 4 48 1
IBS 0 0 0 0 6 0
JPT 0 0 0 1 66 2
LWK 0 0 0 2 66 1
MXL 0 0 0 0 54 0
PUR 0 0 0 1 50 0
TSI 0 0 0 1 81 1
YRI 0 0 0 3 67 1
AFR 0 0 0 5 133 2
AFR+ 0 0 0 6 177 2
AMR 0 0 0 3 147 3
ASN 0 0 0 6 230 2
EUR 0 0 0 10 258 2
ALL 0 0 0 25 812 9
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI