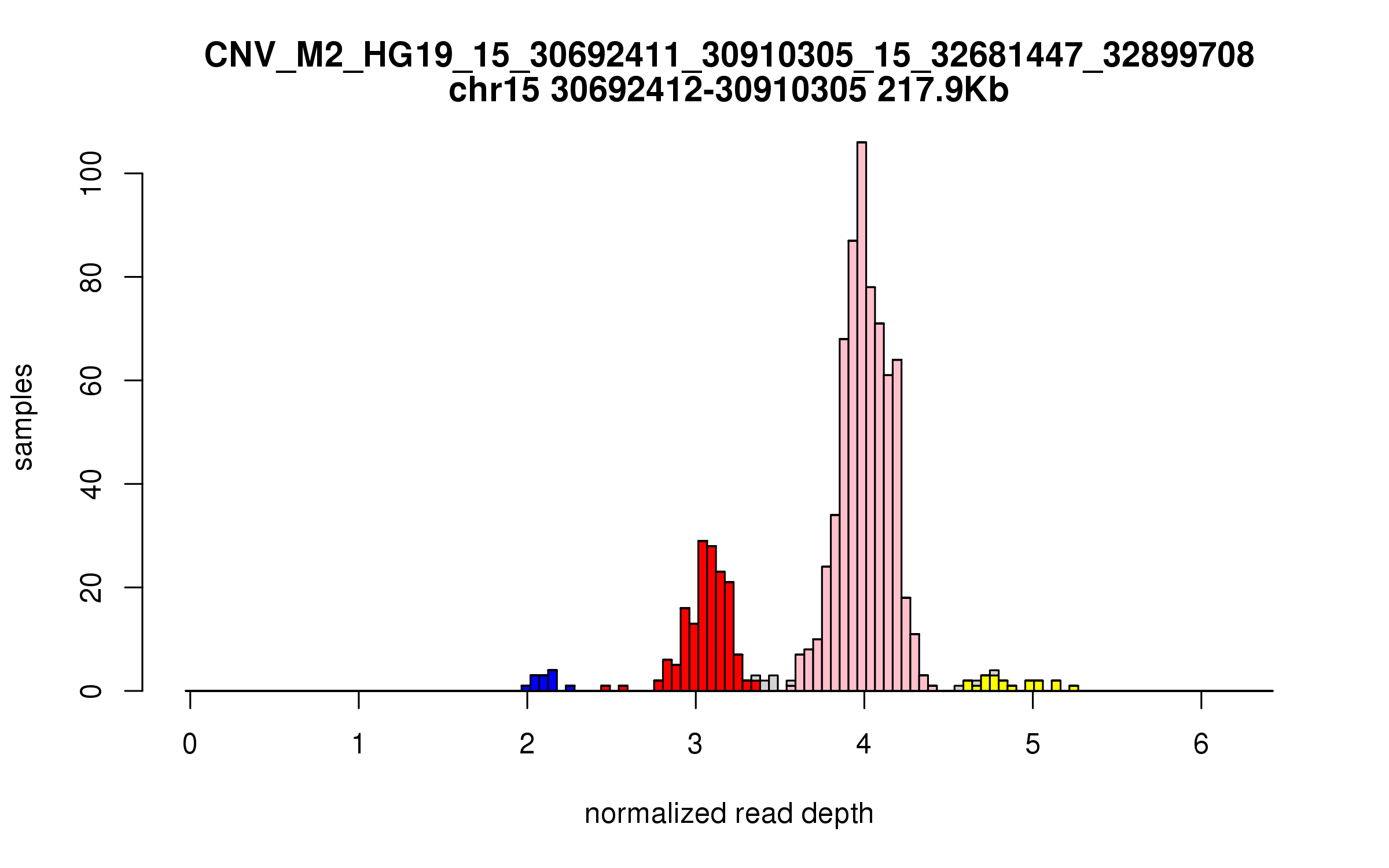
Variant CNV_M2_HG19_15_30692411_30910305_15_32681447_32899708
Location: chr15:30692411-30910305 (217Kb), chr15:32681447-32899708 (218Kb), distance: 1.8Mb
Site classification: Multi-allelic
Variant DCN frequency: 0.223
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.223
Genes Overlapped
| AC135983.2 | GENE | NA |
| AC139426.2 | GENE | NA |
| GOLGA8H | GENE | NA |
| GOLGA8K | GENE | NA |
| GOLGA8N | GENE | NA |
| GOLGA8O | GENE | golgin subfamily A member 8O |
| GOLGA8Q | GENE | NA |
| GOLGA8R | GENE | NA |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 2 11 31 1
CEU 0 0 1 11 50 4
CHB 0 0 1 12 62 3
CHS 0 0 0 10 78 3
CLM 0 0 0 8 40 1
FIN 0 0 0 7 55 0
GBR 0 0 0 11 42 1
IBS 0 0 0 1 5 0
JPT 0 0 1 10 57 1
LWK 0 0 2 30 36 1
MXL 0 0 0 10 42 2
PUR 0 0 1 8 40 3
TSI 0 0 2 10 70 1
YRI 0 0 2 20 49 0
AFR 0 0 4 50 85 1
AFR+ 0 0 6 61 116 2
AMR 0 0 1 26 122 6
ASN 0 0 2 32 197 7
EUR 0 0 3 40 222 6
ALL 0 0 12 159 657 21
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 2 11 31 1
CEU 0 0 1 11 50 4
CHB 0 0 1 12 62 1
CHS 0 0 0 10 78 3
CLM 0 0 0 8 39 1
FIN 0 0 0 7 54 0
GBR 0 0 0 11 42 1
IBS 0 0 0 1 5 0
JPT 0 0 1 10 56 1
LWK 0 0 2 29 35 1
MXL 0 0 0 10 42 2
PUR 0 0 1 8 40 3
TSI 0 0 2 9 69 1
YRI 0 0 2 19 49 0
AFR 0 0 4 48 84 1
AFR+ 0 0 6 59 115 2
AMR 0 0 1 26 121 6
ASN 0 0 2 32 196 5
EUR 0 0 3 39 220 6
ALL 0 0 12 156 652 19
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI