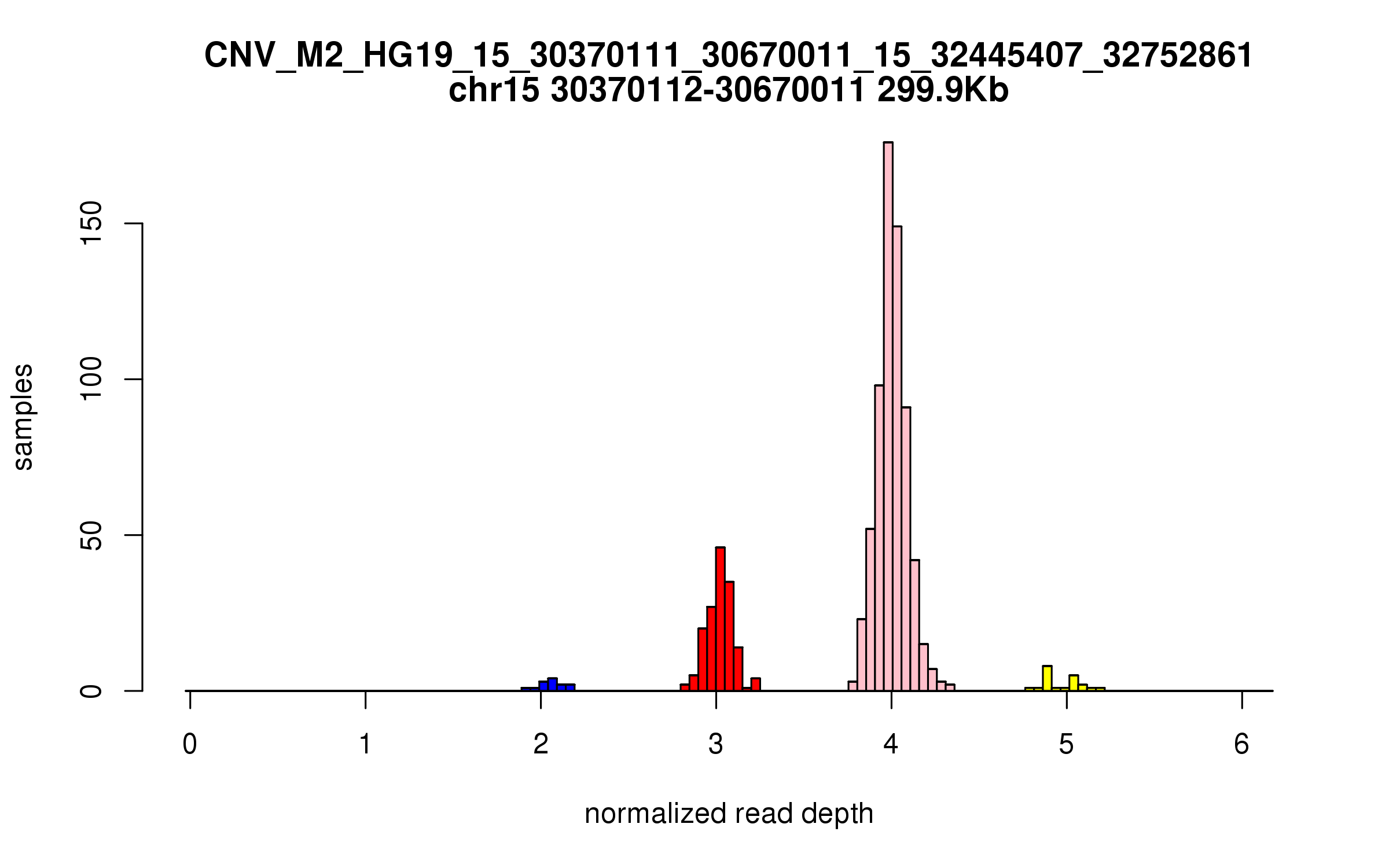
Variant CNV_M2_HG19_15_30370111_30670011_15_32445407_32752861
Location: chr15:30370111-30670011 (299Kb), chr15:32445407-32752861 (307Kb), distance: 1.8Mb
Site classification: Multi-allelic
Variant DCN frequency: 0.221
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.221
Genes Overlapped
| AC139426.2 | GENE | NA |
| CHRFAM7A | CDS | CHRNA7-FAM7A fusion protein isoform 1 |
| CHRNA7 | CDS | NA |
| GOLGA8J | GENE | NA |
| GOLGA8K | GENE | NA |
| GOLGA8O | GENE | golgin subfamily A member 8O |
| GOLGA8T | GENE | NA |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 2 11 31 1
CEU 0 0 1 10 51 4
CHB 0 0 1 12 62 3
CHS 0 0 0 10 78 3
CLM 0 0 0 7 41 1
FIN 0 0 0 7 55 0
GBR 0 0 0 10 43 1
IBS 0 0 0 1 5 0
JPT 0 0 1 10 57 1
LWK 0 0 2 29 37 1
MXL 0 0 0 10 42 2
PUR 0 0 1 8 40 3
TSI 0 0 2 10 70 1
YRI 0 0 3 19 49 0
AFR 0 0 5 48 86 1
AFR+ 0 0 7 59 117 2
AMR 0 0 1 25 123 6
ASN 0 0 2 32 197 7
EUR 0 0 3 38 224 6
ALL 0 0 13 154 661 21
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 2 11 31 1
CEU 0 0 1 10 51 4
CHB 0 0 1 12 62 3
CHS 0 0 0 10 78 3
CLM 0 0 0 7 41 1
FIN 0 0 0 7 55 0
GBR 0 0 0 10 43 1
IBS 0 0 0 1 5 0
JPT 0 0 1 10 57 1
LWK 0 0 2 29 37 1
MXL 0 0 0 10 42 2
PUR 0 0 1 8 40 3
TSI 0 0 2 10 70 1
YRI 0 0 3 19 49 0
AFR 0 0 5 48 86 1
AFR+ 0 0 7 59 117 2
AMR 0 0 1 25 123 6
ASN 0 0 2 32 197 7
EUR 0 0 3 38 224 6
ALL 0 0 13 154 661 21
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI