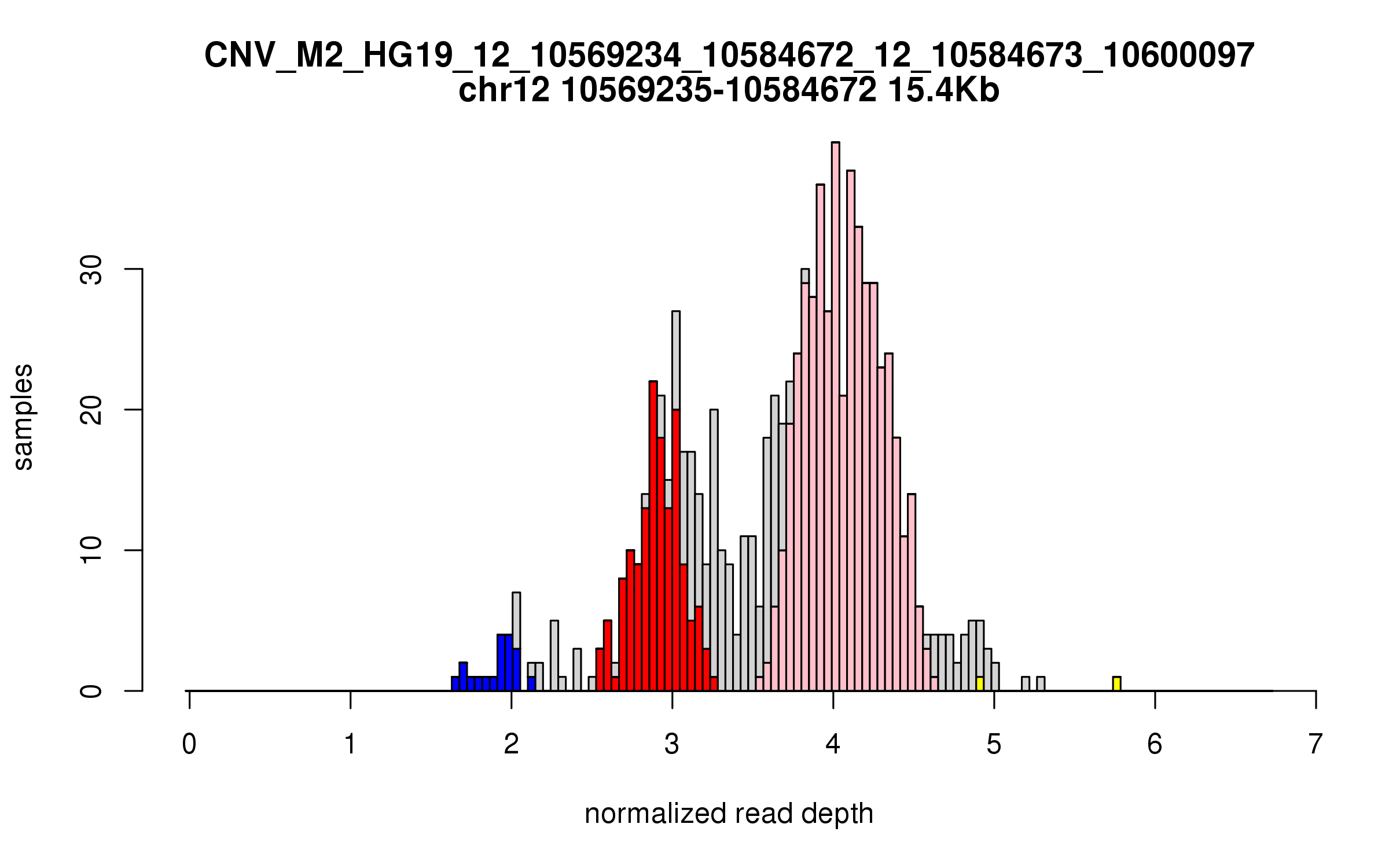
Variant CNV_M2_HG19_12_10569234_10584672_12_10584673_10600097
Location: chr12:10569234-10584672 (15Kb), chr12:10584673-10600097 (15Kb), distance: 0b
Site classification: Multi-allelic
Variant DCN frequency: 0.262
Genes Overlapped
Site classification: Multi-allelic
Variant DCN frequency: 0.262
Genes Overlapped
| KLRC1 | CDS | killer cell lectin-like receptor subfamily C, member 1 |
| KLRC2 | CDS | NKG2-C type II integral membrane protein |
| KLRC3 | CDS | NKG2-E type II integral membrane protein isoform E |
| NKG2-E | CDS | NA |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 3 14 28 0
CEU 0 0 0 23 43 0
CHB 0 0 3 20 55 0
CHS 0 0 4 28 56 3
CLM 0 0 1 8 38 2
FIN 0 0 3 18 41 0
GBR 0 0 1 10 43 0
IBS 0 0 0 1 5 0
JPT 0 0 5 16 46 2
LWK 0 0 1 29 39 0
MXL 0 0 1 13 39 1
PUR 0 0 4 11 37 0
TSI 0 0 2 24 57 0
YRI 0 0 1 24 46 0
AFR 0 0 2 53 85 0
AFR+ 0 0 5 67 113 0
AMR 0 0 6 32 114 3
ASN 0 0 12 64 157 5
EUR 0 0 6 76 189 0
ALL 0 0 29 239 573 8
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2 CN3 CN4 CN5
ASW 0 0 1 8 22 0
CEU 0 0 0 17 35 0
CHB 0 0 2 14 49 0
CHS 0 0 3 12 49 1
CLM 0 0 1 4 30 1
FIN 0 0 2 6 31 0
GBR 0 0 1 4 38 0
IBS 0 0 0 1 3 0
JPT 0 0 4 11 38 0
LWK 0 0 1 20 27 0
MXL 0 0 1 9 31 0
PUR 0 0 2 8 27 0
TSI 0 0 1 12 47 0
YRI 0 0 0 20 43 0
AFR 0 0 1 40 70 0
AFR+ 0 0 2 48 92 0
AMR 0 0 4 21 88 1
ASN 0 0 9 37 136 1
EUR 0 0 4 40 154 0
ALL 0 0 19 146 470 2
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI