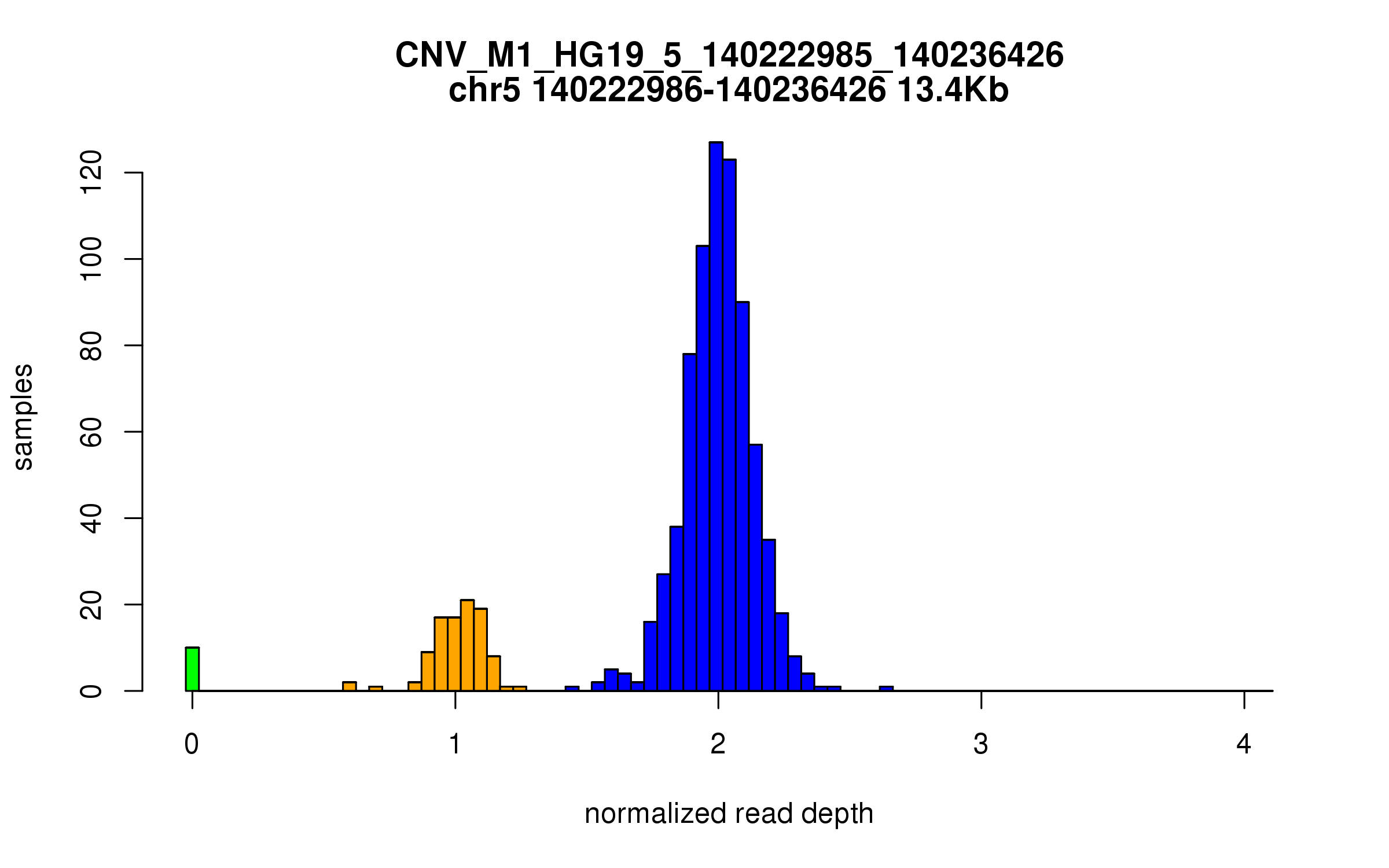
Variant CNV_M1_HG19_5_140222985_140236426
Location: chr5:140222985-140236426 (13Kb)
Site classification: Deletion
Variant DCN frequency: 0.127
Genes Overlapped
Site classification: Deletion
Variant DCN frequency: 0.127
Genes Overlapped
| PCDHA1 | INTRON | protocadherin alpha-1 isoform 1 precursor |
| PCDHA10 | CDS | protocadherin alpha-10 isoform 2 precursor |
| PCDHA2 | INTRON | protocadherin alpha-2 precursor |
| PCDHA3 | INTRON | protocadherin alpha-3 precursor |
| PCDHA4 | INTRON | protocadherin alpha-4 isoform 2 precursor |
| PCDHA5 | INTRON | protocadherin alpha-5 precursor |
| PCDHA6 | INTRON | protocadherin alpha-6 |
| PCDHA7 | INTRON | protocadherin alpha-7 precursor |
| PCDHA8 | CDS | protocadherin alpha-8 precursor |
| PCDHA9 | CDS | protocadherin alpha-9 precursor |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2
ASW 1 5 39
CEU 3 8 55
CHB 1 4 73
CHS 0 1 90
CLM 1 10 38
FIN 0 11 51
GBR 0 9 45
IBS 0 0 6
JPT 1 4 64
LWK 2 14 53
MXL 0 9 45
PUR 0 5 47
TSI 1 9 73
YRI 0 9 62
AFR 2 23 115
AFR+ 3 28 154
AMR 1 24 130
ASN 2 9 227
EUR 4 37 230
ALL 10 98 741
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2
ASW 1 5 39
CEU 3 8 55
CHB 1 4 73
CHS 0 1 90
CLM 1 10 38
FIN 0 11 51
GBR 0 9 45
IBS 0 0 6
JPT 1 4 64
LWK 2 14 53
MXL 0 9 45
PUR 0 5 47
TSI 1 9 73
YRI 0 9 62
AFR 2 23 115
AFR+ 3 28 154
AMR 1 24 130
ASN 2 9 227
EUR 4 37 230
ALL 10 98 741
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI