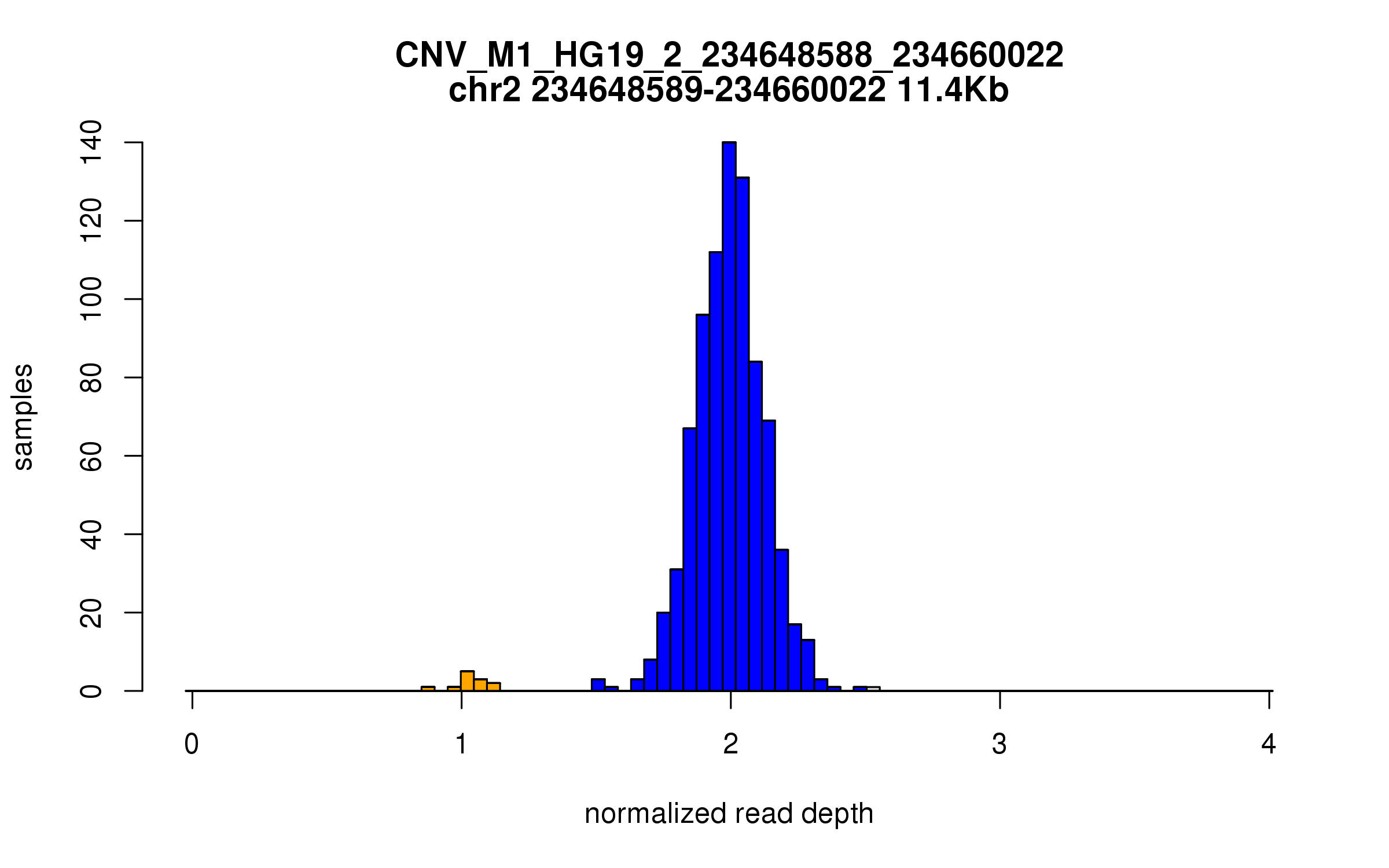
Variant CNV_M1_HG19_2_234648588_234660022
Location: chr2:234648588-234660022 (11Kb)
Site classification: Deletion
Variant DCN frequency: 0.014
Genes Overlapped
Site classification: Deletion
Variant DCN frequency: 0.014
Genes Overlapped
| UGT1A10 | INTRON | UDP-glucuronosyltransferase 1-10 precursor |
| UGT1A3 | INTRON | UDP-glucuronosyltransferase 1-3 precursor |
| UGT1A4 | INTRON | UDP-glucuronosyltransferase 1-4 precursor |
| UGT1A5 | INTRON | UDP-glucuronosyltransferase 1-5 precursor |
| UGT1A6 | INTRON | UDP-glucuronosyltransferase 1-6 precursor |
| UGT1A7 | INTRON | UDP glucuronosyltransferase 1 family, polypeptide A7 precursor |
| UGT1A8 | INTRON | UDP-glucuronosyltransferase 1-8 precursor |
| UGT1A9 | INTRON | UDP-glucuronosyltransferase 1-9 precursor |
1000 Genomes Phase 1
Diploid copy number distribution
POP CN0 CN1 CN2 CN3
ASW 0 0 45 0
CEU 0 0 66 0
CHB 0 0 77 1
CHS 0 0 91 0
CLM 0 1 48 0
FIN 0 0 62 0
GBR 0 0 54 0
IBS 0 0 6 0
JPT 0 0 69 0
LWK 0 7 62 0
MXL 0 0 54 0
PUR 0 0 52 0
TSI 0 0 83 0
YRI 0 4 67 0
AFR 0 11 129 0
AFR+ 0 11 174 0
AMR 0 1 154 0
ASN 0 0 237 1
EUR 0 0 271 0
ALL 0 12 836 1
Diploid copy number distribution (95% confident)
POP CN0 CN1 CN2
ASW 0 0 45
CEU 0 0 66
CHB 0 0 77
CHS 0 0 91
CLM 0 1 48
FIN 0 0 62
GBR 0 0 54
IBS 0 0 6
JPT 0 0 69
LWK 0 7 62
MXL 0 0 54
PUR 0 0 52
TSI 0 0 83
YRI 0 4 67
AFR 0 11 129
AFR+ 0 11 174
AMR 0 1 154
ASN 0 0 237
EUR 0 0 271
ALL 0 12 836
Imputation spider plots: ASW CEU CHB CHS CLM FIN GBR IBS JPT LWK MXL PUR TSI YRI